| Label | Explanation | Data Type |
Input Features
| The input features containing the fields of the two analysis variables. | Feature Layer |
Analysis Field 1
| The field of the first analysis variable. The field must be numeric. | Field |
Analysis Field 2
| The field of the second analysis variable. The field must be numeric. | Field |
Output Features
| The output features containing the local Lee's L statistics, spatial association categories, p-values, and the weighted averages of the neighbors of each feature. | Feature Class |
Neighborhood Type (Optional) | Specifies how neighbors of each feature will be determined. The feature is always included in the neighborhood, and all neighborhood weights are normalized to sum to 1.
| String |
Distance Band
(Optional) | The distance band that will be used to determine neighbors around the focal feature. If no value is provided, the distance will be the shortest distance such that each feature has at least one other neighbor in its neighborhood. For polygons, the distance between centroids will be used to determine neighbors. | Linear Unit |
Number of Neighbors
(Optional) | The number of neighbors around each feature that will be included as neighbors. The value does not include the feature. For example, specifying 6 will use the feature and its six closest neighbors (seven features total). The default is 8. The value must be at least 2. | Long |
Weights Matrix File
(Optional) | The path and file name of the spatial weights matrix file that defines the neighbors and weights between features. | File |
Local Weighting Scheme
(Optional) | Specifies the weighting scheme that will be applied to neighbors when calculating spatial associations.
| String |
Kernel Bandwidth
(Optional) | The bandwidth for the bisquare kernel. The bandwidth defines how quickly the weights decrease with distance. Larger bandwidths will provide comparatively larger weights to neighbors that are farther away from the feature. For the k nearest neighbors neighborhood, the default value (empty) will use an adaptive bandwidth equal to the distance to the (k+1)th neighbor of the focal feature. For the fixed distance band neighborhood, the default (empty) will use the same value as the distance band. | Linear Unit |
Number of Permutations (Optional) | Specifies the number of permutations that will be used to create reference distributions when calculating global and local p-values. All p-values are calculated using two-sided hypothesis tests.
| Long |
Derived Output
| Label | Explanation | Data Type |
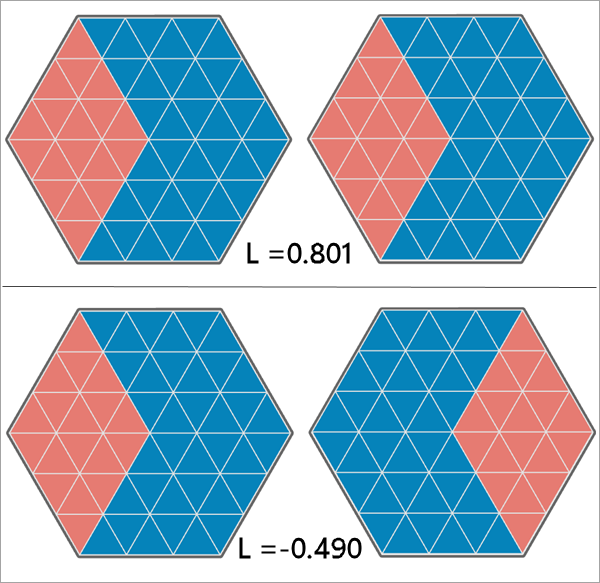
| Lee's L | The Lee's L statistic for the analysis variables. | Double |
| P-value | The p-value for the Lee's L statistic. | Double |
| Pearson Correlation | The Pearson correlation between the analysis variables. | Double |